05/27/2015
Objective: Start DNA extraction on sample "night 2" an additional tissue sample taken during the manipulation experiment. DNA extraction preformed with E.Z.N.A. mollusk dna kit.
Methods: E.Z.N.A.® Mollusc DNA Protocol
Invertebrates preserved in formalin should be rinsed in xylene and then ethanol before processing. Note that results obtained with formalin-fixed tissues generally depend on age and size of specimen. Purified material is usually adequate for PCR amplification, but fresh or frozen samples should be used for Southern analyses.
Materials and Equipment to be Supplied by User: •
Microcentrifuge capable of at least 10,000 x g • Nuclease-free 1.5 mL or 2 mL microcentrifuge tubes • Water baths capable of 60°C and 70°C • Vortexer • 100% ethanol • Chloroform:isoamyl alcohol (24:1)
Before Starting: • Prepare buffers according to the instructions on Page 4 • Set water baths to 60°C and 70°C • Heat Elution Buffer to 70°C 1.
Homogenize tissue sample following one of the procedures below depending on the sample type. A. Arthropods
1. Pulverize no more than 50 mg of tissue in liquid nitrogen with a mortar and pestle. Note: If ceramic mortar and pestle are not available, homogenize the sample in the microcentrifuge tube using a disposable microtube pestle (Omega Bio-tek, Cat No. SSI-1015-39; Eppendorf, Cat No. 0030 120.973; VWR, Cat No. KT 749520- 0000).
2. Transfer the powder to a clean 1.5 mL microcentrifuge tube.
3. Proceed to Step 2 below. E.Z.N.A.® Mollusc DNA Kit Protocol
B. Molluscs (and other soft tissue invertebrates)
1. Pulverize no more than 30 mg tissue in liquid nitrogen with a mortar and pestle.
2. Transfer the powder to a clean 1.5 mL microcentrifuge tube. Note: If ceramic mortar and pestle are not available, homogenize the sample in the microcentrifuge tube using a disposable microtube pestle (Omega Bio-tek, Cat No. SSI-1015-39; Eppendorf, Cat No. 0030 120.973; VWR, Cat No. KT 749520-0000). Addition of a pinch of white quartz sand, -50 to 70 mesh (Sigma Chemical Co. Cat No. S9887) will help.
3. Proceed with Step 2 below.
Note: The amount of starting material depends on the sample and can be increased if acceptable results are obtained with the suggested 30 mg tissue. For easy-to-process specimens, the procedure may be scaled up and the buffer volumes used increased in proportion. In any event, use no more than 50 mg tissue per HiBind® DNA Mini Column as binding capacity (100 μg) may be exceeded. Meanwhile, difficult tissues may require starting with less than 30 mg tissue and doubling all volumes to ensure adequate lysis.
2. Add 350 μL ML1 Buffer and 25 μL Proteinase K Solution. Vortex to mix thoroughly.
3. Incubate at 60°C for a minimum of 30 minutes or until entire sample is solubilized. Note: Actual incubation time varies and depends on the elasticity of the tissue. Most samples require no more than 4 hours. Alternatively an overnight incubation at 37°C will produce adequate results.
4. Add 350 μL chloroform:isoamyl alcohol (24:1). Vortex to mix thoroughly.
5. Centrifuge 10,000 x g for 2 minutes at room temperature.
6. Transfer the upper aqueous phase to a clean 1.5 mL microcentrifuge tube. Avoid the milky interface containing contaminants and inhibitors. Note: This step will remove much of the polysaccharides and proteins from solution and improve spin-column performance downstream. If a small upper aqueous phase is present after centrifugation, add 200 μL ML1 Buffer and vortex to mix thoroughly. Repeat Step 5 (centrifugation) and Step 6 (transfer the upper aqueous phase).
7. Add one volume MBL Buffer and 10 μL RNase A. Vortex at maximum speed for 15 seconds. Note: For example, to 500 μL upper aqueous solution from Step 6, add 500 μL MBL Buffer.
8. Incubate at 70°C for 10 minutes.
9. Cool the sample to room temperature.
10. Add one volume 100% ethanol. Vortex at maximum speed for 15 seconds. Note: For example, to 500 μL upper aqueous solution from Step 6, add 500 μL 100% ethanol.
11. Insert a HiBind® DNA Mini Column into a 2 mL Collection Tube. Optional Protocol for Column Equilibration: 1. Add 100 µL 3M NaOH to the HiBind® DNA Mini Column. 2. Centrifuge at maximum speed for 60 seconds. 3. Discard the filtrate and reuse the collection tube.
12. Transfer 750 µL sample from Step 9 (including any precipitate that may have formed) to the HiBind® DNA Mini Column.
13. Centrifuge at 10,000 x g for 1 minute.
14. Discard the filtrate and reuse the collection tube.
15. Repeat Steps 12-14 until all of the sample has been applied to the HiBind® DNA Mini Column.
16. Discard the filtrate and the Collection Tube.
17. Insert the HiBind® DNA Mini Column into a new 2 mL Collection Tube.
18. Add 500 μL HBC Buffer. Note: HBC Buffer must be diluted with isopropanol before use. Please see Page 6 for instructions.
19. Centrifuge at 10,000 x g for 30 seconds.
20. Discard the filtrate and reuse the Collection Tube.
21. Add 700 μL DNA Wash Buffer. Note: DNA Wash Buffer must be diluted with 100% ethanol prior to use. Please refer to Page 4 or the bottle label for instructions. 22. Centrifuge at 10,000 x g for 1 minute.
23. Discard the filtrate and reuse the Collection Tube.
24. Repeat Steps 21-235 for a second DNA Wash Buffer wash step.
25. Centrifuge the empty HiBind® DNA Mini Column at maximum speed for 2 minutes to dry the membrane.
Note: It is critical to remove any trace of ethanol that may otherwise interfere with downstream applications.
26. Transfer the HiBind® DNA Mini Column to a nuclease-free 1.5 or 2 mL microcentrifuge tube (not provided).
27. Add 50-100 μL Elution Buffer (or sterile deionized water) preheated to 70°C. Note: Smaller elution volumes will increase DNA concentration but decrease yield. Elution volumes greater than 200 μL are not recommended.
28. Let sit at room temperature for 2 minutes.
29. Centrifuge at 10,000 x g for 1 minute.
30. Repeat Steps 27-29 for a second elution step. Note: Any combination of the following steps can be used to help increase DNA yield. • After adding the Elution Buffer, incubate the column for 5 minutes. • Increase the elution volume. • Repeat the elution step with fresh Elution Buffer (this may increase the yield, but decrease the concentration). • Repeat the elution step using the eluate from the first elution (this may increase yield while maintaining elution volume).
31. Store DNA at -20°C
Results: Successfully extracted DNA from tissues will now use PCR to replicate extraction product
Wednesday, May 27, 2015
Tuesday, May 26, 2015
05/26/2015
Objective: To run PCR product on a gel with same measurements and ladder as 05/18/2015, lane 1 and 2 loaded with P. gurneyi cDNA and lane 4 were controls loaded with H20 instead of template.
Results: Was expecting a product size of 822 but the gel shows around 200. additionally the bands are not well defined. No contamination seen in the gel will continue on 05/27/2015 with DNA extraction using tissue from the manipulation experiment using a partial sample to keep some for later experiment.
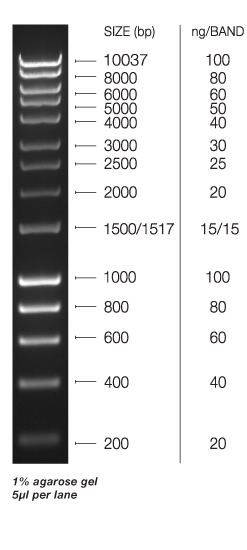

Objective: To run PCR product on a gel with same measurements and ladder as 05/18/2015, lane 1 and 2 loaded with P. gurneyi cDNA and lane 4 were controls loaded with H20 instead of template.
Results: Was expecting a product size of 822 but the gel shows around 200. additionally the bands are not well defined. No contamination seen in the gel will continue on 05/27/2015 with DNA extraction using tissue from the manipulation experiment using a partial sample to keep some for later experiment.

Wednesday, May 20, 2015
Monday, May 18, 2015
05/18/2015
Objective: run PCR products in Friedman lab from 05/13/2015 on a gel
50ml 1x TBE
.5 grams agarose (1%)
5 ul athidium bromide
Gel run with hyper ladder I


Went over methods and materials with Sam all that's left is to hear back from Jake and will be able to finish up proposal
Results: verified that reagents had contamination. The experiment should of not shown any bands because template cDNA was replaced with H2O. On Wednesday will start with new reagents.
Gel
Objective: run PCR products in Friedman lab from 05/13/2015 on a gel
50ml 1x TBE
.5 grams agarose (1%)
5 ul athidium bromide
Gel run with hyper ladder I

Went over methods and materials with Sam all that's left is to hear back from Jake and will be able to finish up proposal
Results: verified that reagents had contamination. The experiment should of not shown any bands because template cDNA was replaced with H2O. On Wednesday will start with new reagents.
Gel
Wednesday, May 13, 2015
05/13/2015
Objective: Start a PCR to try to figure out contamination from 04/23.
Results: Ran PCR with H20 instead of template following all the measurements and running times of 04/22/2015. Will run gel on Monday with Sam and hopefully start another run with template to get ready for cloning.
Spent the remaining time working on finishing up the proposal. Sent off to Sam to verify methods and materials, and Jake to proof read. Updated intro, work cited, schedule, abstract.
Objective: Start a PCR to try to figure out contamination from 04/23.
Results: Ran PCR with H20 instead of template following all the measurements and running times of 04/22/2015. Will run gel on Monday with Sam and hopefully start another run with template to get ready for cloning.
Spent the remaining time working on finishing up the proposal. Sent off to Sam to verify methods and materials, and Jake to proof read. Updated intro, work cited, schedule, abstract.
Subscribe to:
Posts (Atom)