Objective: Since the sequencing data that was collected did not fit the luciferase sequencing obtained from Renilla reniformes, we are running another. Started a PCR using all the measurements from 4/22/2015 using only the luciferase primers. Through Further research I did find multiple studies involving the bacteria and luciferase, though I am not sure if that has a connection to the results
Liu, Y., Abaibou, H., & Fletcher, H. M. (2000). Development of a noninvasive reporter system for gene expression in Porphyromonas gingivalis. Plasmid,44(3), 250-261.
Coats, S. R., Reife, R. A., Bainbridge, B. W., Pham, T. T. T., & Darveau, R. P. (2003). Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at toll-like receptor 4 in human endothelial cells.Infection and immunity, 71(12), 6799-6807.
1 RR Luciferase
2 RR Luciferase
3 H20 Control
4 H20 Control
Dr. Roberts suggested blasting each sequence
Results: Sucessfully prepared a gel and started the PCR. Spent the remaining time working with Sam to go over Sequencing data.
Geneious work
Worked with sam to go over sequencing data using geneious 5.3.6 trimmed all 12 sequences, aligned and then assembled shown below. The red areas are the areas that were trimmed leaving the un-red areas to be aligned. Blasted the sequence that was created in NCBI using nucleotide blast, tbalstx (compares proteins), and Compared directly to the sequence that was used to prepare primers. Both blasts did not return anything related to Luciferase or Renilla Reniformes. Since this sequence has never been sequenced before may be why nothing is relating. The next PCR is completely focused on luciferase which gave us good results last run through. I am planning on going through and blasting each sequence at a later date. Sam and I both think there may be something going on since the sequencing results are a lot shorter than what was being sent out. The PCR bands were over 800 base pairs but all the sequencing data has been between 200 to 300 bp.
Residues:
(-------------------------------CGGCTCTGCCTCGCGTATCGATAT-GATTTGGTTGCCTACCCGCTTTTCCGGGTTTATTAATAAAGTWRAAATCTTACCATGTTTCCGGTCGGACCATCATTATGTGTTTATTGAGATGCATTTACCTTTTTCTGTTGTTCGCGGCAATGAGCTTTGGAAACTCAATTTTTCATTATTAAAAGACGAATGTTTATGCCAGAAAATTATGGACTTCTGGAAGATATGGAAGGTCCAAAAACATGTTTACCTTCCCTCCGTTTGGTGGGAACTTGGAAAAAAGCGTCTTATCGACATTATCCGCCGTTTCGGTCGAAGCCKTGCTAGCGCAGTGCGCGATCGTGTTGCTGATTTAACTGCCCAATTGAATGCATGCAAAA-----------------)
Went into the literature to reverify the presence of luciferase in ptilosarcus gurneyi did not find much information only one study (shown below)
Shimomura, O., & Johnson, F. H. (1979). Comparison of the amounts of key components in the bioluminescence systems of various coelenterates.Comparative Biochemistry and Physiology Part B: Comparative Biochemistry,64(1), 105-107.
Monday, June 29, 2015
Thursday, June 25, 2015
Blasted Sequence
Objective: Blast the sequence on NCBI to see if it matches any known sequences.
Results: found a connection to a bacteria (shown below)
Results: found a connection to a bacteria (shown below)
Wednesday, June 24, 2015
Sequencing information
Objective: Work with Geneious 5.6.7 basic addition to align sequencing data. Take Data from multiple PCRs sent out for sequencing cDNA and DNA
Link to data http://eagle.fish.washington.edu/scaphapoda/index.php?dir=Jonathan%2F
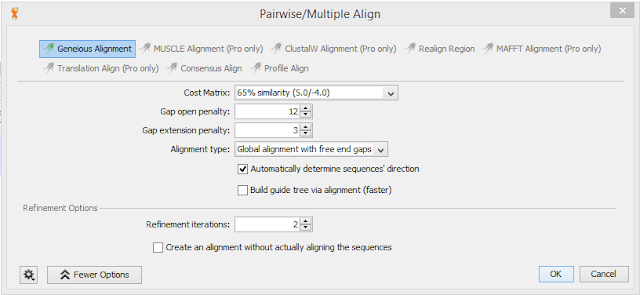
Results: Results from sequenced multiple pieces though still quite a few ambiguities. Included settings used in Geneious for reference. I will need to go over results with Dr. Roberts and Sam to interpret and possible redo alignment for better results. Tried multiple Cost Matrix with varied results so I was not sure if I was using the program correctly.
Link to data http://eagle.fish.washington.edu/scaphapoda/index.php?dir=Jonathan%2F
Results: Results from sequenced multiple pieces though still quite a few ambiguities. Included settings used in Geneious for reference. I will need to go over results with Dr. Roberts and Sam to interpret and possible redo alignment for better results. Tried multiple Cost Matrix with varied results so I was not sure if I was using the program correctly.
Wednesday, June 17, 2015
06/17/2015
Objective: Started cleaning up notebook go through and start adding in "Objective" and "Results" for each work day. Fill out any missing information. Decided to start using Blogger again because of user friendliness and to transfer into wiki. (http://genefish.wikispaces.com/Jonathan%27s+Notebook)
Results: Updated large portion of the posts and added comments and additional information. A good start to cleaning up the notebook need to sit down with Sam and make sure all necessary details are in place.
Objective: Started cleaning up notebook go through and start adding in "Objective" and "Results" for each work day. Fill out any missing information. Decided to start using Blogger again because of user friendliness and to transfer into wiki. (http://genefish.wikispaces.com/Jonathan%27s+Notebook)
Results: Updated large portion of the posts and added comments and additional information. A good start to cleaning up the notebook need to sit down with Sam and make sure all necessary details are in place.
Thursday, June 4, 2015
06/04/2015
Objective: Run PCR products from 06/01/2015 on a gel to verify Luciferase and GFP primers were correct, and if primers were correct and bands are shown remove bands and send out for sequencing
Ran out PCR products from 06/01/2015 on gel
Lanes as seen in the picture,
1 Ladder (Hyper Ladder 1)
2 GFP 1 (P1)
3 GFP 2 (P2)
4 Control
5 Control
6 Blank
7 Blank
10 Control
11 Control
Results: Lane 2 and 3 showed multiple bands none within anticipated length of 1054 but still removed brightest band in lane 3 in line with size "800" on the ladder. Lane 8 and 9 showed bright bands that were in line with the correct size of 822 removed both bands. All removed bands were sent purified and sent out for sequencing by Sam
Objective: Run PCR products from 06/01/2015 on a gel to verify Luciferase and GFP primers were correct, and if primers were correct and bands are shown remove bands and send out for sequencing
Ran out PCR products from 06/01/2015 on gel
Lanes as seen in the picture,
1 Ladder (Hyper Ladder 1)
2 GFP 1 (P1)
3 GFP 2 (P2)
4 Control
5 Control
6 Blank
7 Blank
8 Luciferase 1 (R1)
9 Luciferase 2 (R2)10 Control
11 Control
12 ladder (Hypper ladder 1)
| sr_ID | Primer name | Primer Sequence | date ordered | #bp | GC% | Organism | Gene | Gene Accession# | location on reference | pair w/sr_ID | product size | IDT # |
| 1604 | Rr_46_65F | CGGATGATAACTGGTCCGCA | 8/20/14 | 20 | 55 | Renilla reniformis | Luciferase | M63501 | 46-65 | 1603 | 822 | 124903282 |
| 1603 | Rr_887_868R | TCAGGTGCATCTTCTTGCGA | 8/20/14 | 20 | 50 | Renilla reniformis | Luciferase | M63501 | 887-868 | 1604 | 822 | 124903283 |
| 1602 | Pg_1_PP6_53_72F | CGGCAAAAGCTAGCGTTGAA | 8/18/14 | 20 | 50 | Ptilosarcus gurneyi | Ptilosarcus Green Fluorescent Protein (GFP) | HV097611 | 53-72 | 1601 | 1054 | 124817505 |
| 1601 | Pg_2_PP9_1107_1088R | ACGTGCGGTCTCTTTATGCT | 8/18/14 | 20 | 50 | Ptilosarcus gurneyi | Ptilosarcus Green Fluorescent Protein (GFP) | HV097611 | 1107-1088 | 1602 | 1054 | 124817506 |
Results: Lane 2 and 3 showed multiple bands none within anticipated length of 1054 but still removed brightest band in lane 3 in line with size "800" on the ladder. Lane 8 and 9 showed bright bands that were in line with the correct size of 822 removed both bands. All removed bands were sent purified and sent out for sequencing by Sam
Monday, June 1, 2015
06/01/2015
Objective: Start a pcr with both luciferase primers, and Green florescent Protein primers with DNA produced on 05/27. Used the same procedures and measurements for the PCR as 04/22/2015 though used different temperatures increasing the 72 degree step from 1 min to 3 min to account for the larger strand size in DNA . Prepared a gel for Wednesday using previous measurements from 5/18/2015.
PCR tubes
Green Florescent Protein PCR P1, P2 (Ptilosarcus gurneyi) template P3, P4 (Water Control)
Luciferase PCR R1, R2 (Renilla reniformes template) R3, R4 (Water Control)
Results: Completed PCR and will run out on prepared gel.
Objective: Start a pcr with both luciferase primers, and Green florescent Protein primers with DNA produced on 05/27. Used the same procedures and measurements for the PCR as 04/22/2015 though used different temperatures increasing the 72 degree step from 1 min to 3 min to account for the larger strand size in DNA . Prepared a gel for Wednesday using previous measurements from 5/18/2015.
PCR tubes
Green Florescent Protein PCR P1, P2 (Ptilosarcus gurneyi) template P3, P4 (Water Control)
Luciferase PCR R1, R2 (Renilla reniformes template) R3, R4 (Water Control)
Results: Completed PCR and will run out on prepared gel.
Subscribe to:
Posts (Atom)